A pair of genuine interacting proteins are generally expected to have a common cellular role and proteins that have common interaction partners have a high chance of sharing a common function. In AtPIN, due to its integrative profile, the reliability index for a reported PPI can be postulated in terms of the proportion of interaction partners that two proteins have in common. The FSWeight implemented in AtPIN was originally proposed by Chua et al?.
The FSWeight is calculated as
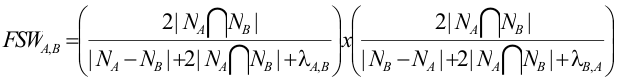
Where,
= Set of interaction partners shared by A and B;
To evaluate one PPI in AtPIN, the C? and FSWeight must be consider together since, the effectiveness of using FSWeight as a PPI reliability index was demonstrated by Wong ?. Using 19.452 interactions in yeast obtained from the GRID database , over 80% of the top 10% of protein interactions ranked by FSWeight have a common cellular role and over 90% of them have a common subcellular localization.
? Chua HN, Sung WK, Wong L: Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics 2006, 22:1623-1630
? Chen J, Hsu W, Lee ML, Ng SK: Increasing confidence of protein interactomes using network topological metrics. Bioinformatics 2006, 22:1998-2004.
Chua HN, Sung WK, Wong L: Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics (Oxford, England) 2006, 22:1623-1630
?